Uncover novel drug targets and resistance mechanisms with genome-scale precision
Uncover novel drug targets and resistance mechanisms with genome-scale precision
CRISPR Functional Genomics Screening
DuneX Biosciences provides comprehensive CRISPR screening services to identify gene dependencies, synthetic lethal interactions, and phenotypic drivers of drug response. Our platform integrates genome-wide knockout or activation libraries with high-content readouts and next-generation sequencing.
Workflow Overview
We handle the complete process — from construct design through assay-ready, cryopreserved vials.

Step 1 — Library Design & Construction
Genome-wide or focused sgRNA libraries (10⁵–10⁶ guides) built for knockout, CRISPRi, or CRISPRa screens.

Step 2 — Cell Line Engineering
Lentiviral transduction or stable Cas9/Cas12a cell line generation with optimized MOI and selection.
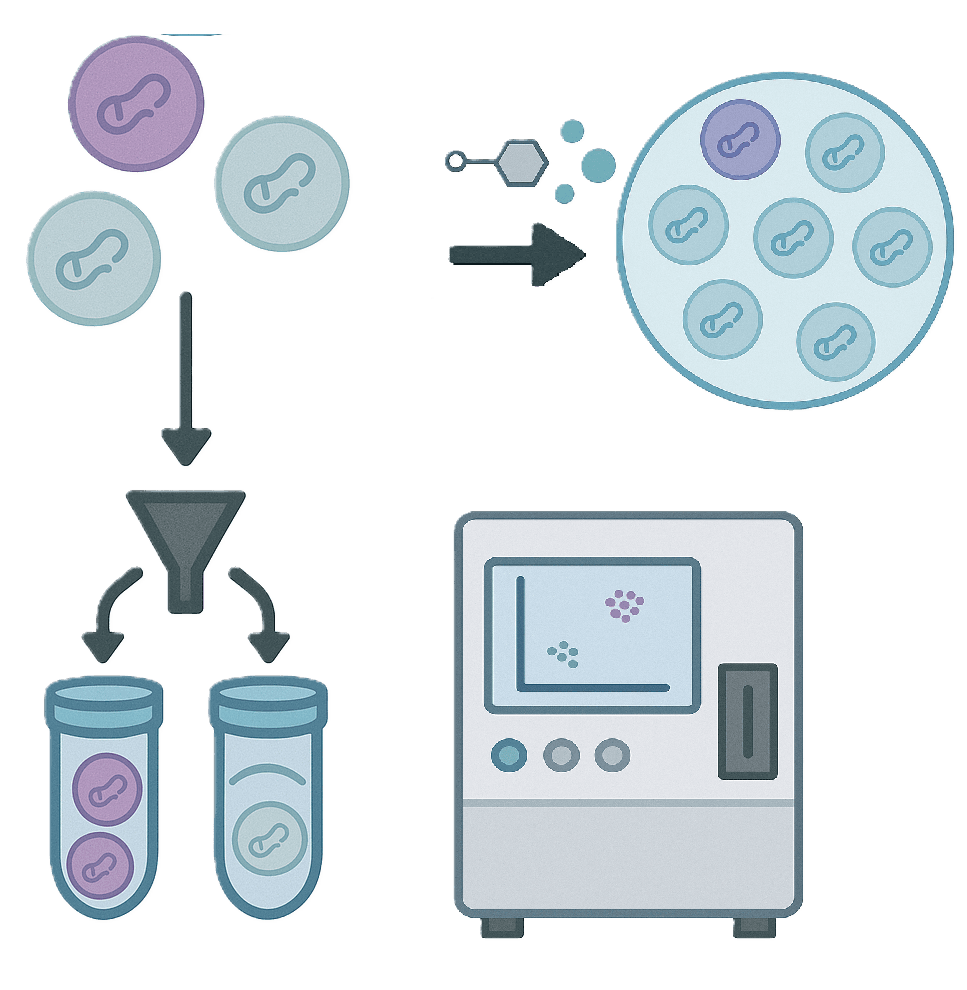
Step 3 — Screening Execution
Drug-treated vs. control, proliferation-based, or FACS-based phenotypic enrichment workflows.

Step 4 — Genomic DNA Extraction
Isolate high-quality genomic DNA from input and selected cell populations.

Step 5 — NGS & Quantification
Amplify sgRNA cassettes for Illumina sequencing and quantify guide abundance.

Step 6 — Hit Analysis & Interpretation
MAGeCK / CRISPResso2 pipeline to identify gene dependencies, resistance mechanisms, and key drivers.
Package Pricing
Drug Synthetic Lethal Screen
Identify genes whose loss enhances or suppresses sensitivity to a specific therapeutic compound. This route is ideal for target identification, mechanism-of-action elucidation, and drug combination discovery.
$
5,000
/sample
**
Genome-wide or custom sgRNA library (10⁵–10⁶ guides)
Drug-treated vs. control cell populations
NGS-based dropout or enrichment analysis (MAGeCK / CRISPResso2)
Hit prioritization and pathway enrichment mapping
** Minimum 2 Samples: Drug treated vs untreated
💡 Deliverables: ranked gene lists, volcano plots, pathway analyses, and validation-ready hit data.
FACS-Based Phenotypic Screen
Discover genetic drivers of surface marker expression, signaling activity, or cell fate using fluorescence-activated cell sorting (FACS) as the selection readout. Ideal for immunology, oncology, and cell therapy programs.
$
7,000
/sample
***
CRISPR knockout / activation libraries in mammalian cells
Sorting based on fluorescent reporter intensity or antibody staining
Sorted population sequencing and differential sgRNA enrichment
Data integration with single-cell expression or pathway analysis
*** Minimum 2 Samples: Low expression vs high expression
🔬 Deliverables: FACS gating data, differential gene lists, and pathway enrichment reports.
Drug Synthetic Lethal Screen
Identify genes whose loss enhances or suppresses sensitivity to a specific therapeutic compound. This route is ideal for target identification, mechanism-of-action elucidation, and drug combination discovery.
$
5,000
/sample
**
Genome-wide or custom sgRNA library (10⁵–10⁶ guides)
Drug-treated vs. control cell populations
NGS-based dropout or enrichment analysis (MAGeCK / CRISPResso2)
Hit prioritization and pathway enrichment mapping
** Minimum 2 Samples: Drug treated vs untreated
💡 Deliverables: ranked gene lists, volcano plots, pathway analyses, and validation-ready hit data.
FACS-Based Phenotypic Screen
Discover genetic drivers of surface marker expression, signaling activity, or cell fate using fluorescence-activated cell sorting (FACS) as the selection readout. Ideal for immunology, oncology, and cell therapy programs.
$
7,000
/sample
***
CRISPR knockout / activation libraries in mammalian cells
Sorting based on fluorescent reporter intensity or antibody staining
Sorted population sequencing and differential sgRNA enrichment
Data integration with single-cell expression or pathway analysis
*** Minimum 2 Samples: Low expression vs high expression
🔬 Deliverables: FACS gating data, differential gene lists, and pathway enrichment reports.
Drug Synthetic Lethal Screen
Identify genes whose loss enhances or suppresses sensitivity to a specific therapeutic compound. This route is ideal for target identification, mechanism-of-action elucidation, and drug combination discovery.
$
5,000
/sample
**
Genome-wide or custom sgRNA library (10⁵–10⁶ guides)
Drug-treated vs. control cell populations
NGS-based dropout or enrichment analysis (MAGeCK / CRISPResso2)
Hit prioritization and pathway enrichment mapping
** Minimum 2 Samples: Drug treated vs untreated
💡 Deliverables: ranked gene lists, volcano plots, pathway analyses, and validation-ready hit data.
FACS-Based Phenotypic Screen
Discover genetic drivers of surface marker expression, signaling activity, or cell fate using fluorescence-activated cell sorting (FACS) as the selection readout. Ideal for immunology, oncology, and cell therapy programs.
$
7,000
/sample
***
CRISPR knockout / activation libraries in mammalian cells
Sorting based on fluorescent reporter intensity or antibody staining
Sorted population sequencing and differential sgRNA enrichment
Data integration with single-cell expression or pathway analysis
*** Minimum 2 Samples: Low expression vs high expression
🔬 Deliverables: FACS gating data, differential gene lists, and pathway enrichment reports.
FAQ
1. What types of CRISPR pooled screens do you support?
1. What types of CRISPR pooled screens do you support?
1. What types of CRISPR pooled screens do you support?
1. What types of CRISPR pooled screens do you support?
2. What are the recommended experimental designs for drug–gene interaction screens?
2. What are the recommended experimental designs for drug–gene interaction screens?
2. What are the recommended experimental designs for drug–gene interaction screens?
2. What are the recommended experimental designs for drug–gene interaction screens?
3. What multiplicity of infection (MOI) do you use for pooled libraries?
3. What multiplicity of infection (MOI) do you use for pooled libraries?
3. What multiplicity of infection (MOI) do you use for pooled libraries?
3. What multiplicity of infection (MOI) do you use for pooled libraries?
4. What sgRNA expression systems and promoters do you support?
4. What sgRNA expression systems and promoters do you support?
4. What sgRNA expression systems and promoters do you support?
4. What sgRNA expression systems and promoters do you support?
5. How do you ensure consistent Cas9 or dCas9 expression across cell populations?
5. How do you ensure consistent Cas9 or dCas9 expression across cell populations?
5. How do you ensure consistent Cas9 or dCas9 expression across cell populations?
5. How do you ensure consistent Cas9 or dCas9 expression across cell populations?
6. What are typical cell numbers and coverage requirements?
6. What are typical cell numbers and coverage requirements?
6. What are typical cell numbers and coverage requirements?
6. What are typical cell numbers and coverage requirements?
7. Can you perform screens in primary or difficult-to-transduce cells?
7. Can you perform screens in primary or difficult-to-transduce cells?
7. Can you perform screens in primary or difficult-to-transduce cells?
7. Can you perform screens in primary or difficult-to-transduce cells?
8. What libraries do you offer?
8. What libraries do you offer?
8. What libraries do you offer?
8. What libraries do you offer?
9. How do you validate library representation prior to screening?
9. How do you validate library representation prior to screening?
9. How do you validate library representation prior to screening?
9. How do you validate library representation prior to screening?
10. Do you support dual-sgRNA or barcoded libraries?
10. Do you support dual-sgRNA or barcoded libraries?
10. Do you support dual-sgRNA or barcoded libraries?
10. Do you support dual-sgRNA or barcoded libraries?
11. What selection strategies do you support?
11. What selection strategies do you support?
11. What selection strategies do you support?
11. What selection strategies do you support?
12. How do you maintain guide representation throughout culture?
12. How do you maintain guide representation throughout culture?
12. How do you maintain guide representation throughout culture?
12. How do you maintain guide representation throughout culture?
13. What controls are used in CRISPR screens?
13. What controls are used in CRISPR screens?
13. What controls are used in CRISPR screens?
13. What controls are used in CRISPR screens?
14. Can you run screens in co-culture or immune-oncology models?
14. Can you run screens in co-culture or immune-oncology models?
14. Can you run screens in co-culture or immune-oncology models?
14. Can you run screens in co-culture or immune-oncology models?
15. What sequencing depth is required?
15. What sequencing depth is required?
15. What sequencing depth is required?
15. What sequencing depth is required?
16. What computational pipelines do you use for hit analysis?
16. What computational pipelines do you use for hit analysis?
16. What computational pipelines do you use for hit analysis?
16. What computational pipelines do you use for hit analysis?
17. What QC metrics do you provide post-screen?
17. What QC metrics do you provide post-screen?
17. What QC metrics do you provide post-screen?
17. What QC metrics do you provide post-screen?
18. Do you provide hit validation and follow-up assays?
18. Do you provide hit validation and follow-up assays?
18. Do you provide hit validation and follow-up assays?
18. Do you provide hit validation and follow-up assays?
19. What are the total timelines for a full screen?
19. What are the total timelines for a full screen?
19. What are the total timelines for a full screen?
19. What are the total timelines for a full screen?
20. What deliverables will I receive?
20. What deliverables will I receive?
20. What deliverables will I receive?
20. What deliverables will I receive?

Copyright © 2025 DuneX Biosciences. All rights reserved. | +1-(415).463.0365 | info@dunexbio.com | 25801 Industrial Blvd Suite 100, Hayward, CA 94545

Copyright © 2025 DuneX Biosciences.
All rights reserved.
+1-(415).463.0365 | info@dunexbio.com |
25801 Industrial Blvd Suite 100, Hayward, CA 94545

Copyright © 2025 DuneX Biosciences. All rights reserved. | +1-(415).463.0365 | info@dunexbio.com |
25801 Industrial Blvd Suite 100, Hayward, CA 94545
